Dynamic Profiling and Prediction of Antibody Response to SARS-CoV-2 Booster-Inactivated Vaccines by Microsample-Driven Biosensor and Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
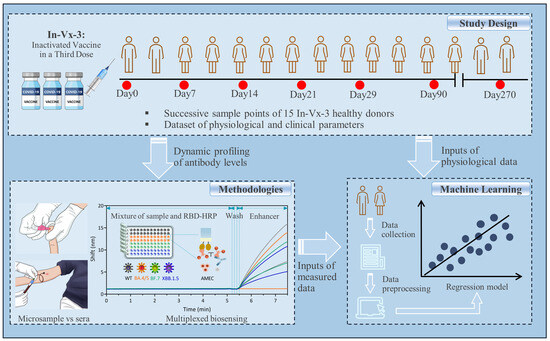
2.1. Study Design
2.2. Materials and Equipment
2.3. Designing the FO-BLI Biosensor for Multiplexed Detection of NAbs
2.4. Evaluating the Fibers’ Reproducibility for FO-BLI NAbs Biosensor
2.5. Establishing DBS Method for NAbs Evaluation
2.6. Description of the Study Cohort
2.7. Designing the FO-BLI Biosensor for Multiplexed Detection of BAbs in Sera
2.8. Defining the Limit of Detection in Both Sera and DBS Samples for NAbs
2.9. Validation of the FO-BLI NAbs Biosensor Using In Vitro PVNT
2.10. Machine Learning-Based Prediction of the Antibody Levels
2.11. Statistical Analysis
3. Results
3.1. Multiplexed FO-BLI Biosensor Ability to Detect NAbs in Sera and Microsamples
3.2. Baseline Characteristics of the Study Cohort
3.3. Dynamic Profiling of WT-NAbs in Both Sera and Microsamples
3.4. Multiplexed Profiling of NAbs towards Omicron BA.4/5, BF.7 and XBB.1.5
3.5. Validation of the FO-BLI NAbs Biosensor Using Clinically Validated PVNT
3.6. Multiplexed Biosensing of Sera BAbs towards Both WT and Variants
3.7. Machine Learning-Based Modeling to Predict Antibody Levels over Time
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ye, Y. China’s rolling COVID waves could hit every six months—Infecting millions. Nature 2023, 618, 442–443. [Google Scholar] [CrossRef]
- Zhou, T.; Shi, T.; Li, A.; Zhu, L.; Zhao, X.; Mao, N.; Qin, W.; Bi, H.; Yang, M.; Dai, M.; et al. A third dose of inactivated SARS-CoV-2 vaccine induces robust antibody responses in people with inadequate response to two-dose vaccination. Natl. Sci. Rev. 2022, 9, nwac066. [Google Scholar] [CrossRef] [PubMed]
- Zeng, G.; Wu, Q.; Pan, H.; Li, M.; Yang, J.; Wang, L.; Wu, Z.; Jiang, D.; Deng, X.; Chu, K.; et al. Immunogenicity and safety of a third dose of CoronaVac, and immune persistence of a two-dose schedule, in healthy adults: Interim results from two single-centre, double-blind, randomised, placebo-controlled phase 2 clinical trials. Lancet Infect. Dis. 2022, 22, 483–495. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P.; et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Khoury, D.S.; Wheatley, A.K.; Ramuta, M.D.; Reynaldi, A.; Cromer, D.; Subbarao, K.; O’Connor, D.H.; Kent, S.J. Measuring immunity to SARS-CoV-2 infection: Comparing assays and animal models. Nat. Rev. Immunol. 2020, 20, 727–738. [Google Scholar] [CrossRef]
- Bian, S.; Shang, M.; Sawan, M. Rapid biosensing SARS-CoV-2 antibodies in vaccinated healthy donors. Biosens. Bioelectron. 2022, 204, 114054. [Google Scholar] [CrossRef]
- Morley, G.L.; Taylor, S.; Jossi, S.; Perez-Toledo, M.; Faustini, S.E.; Marcial-Juarez, E.; Shields, A.M.; Goodall, M.; Allen, J.D.; Watanabe, Y.; et al. Sensitive Detection of SARS-CoV-2-Specific Antibodies in Dried Blood Spot Samples. Emerg. Infect. Dis. 2020, 26, 2970–2973. [Google Scholar] [CrossRef] [PubMed]
- Mulchandani, R.; Brown, B.; Brooks, T.; Semper, A.; Machin, N.; Linley, E.; Borrow, R.; Wyllie, D.; EDSAB-HOME Study Investigators. Use of dried blood spot samples for SARS-CoV-2 antibody detection using the Roche Elecsys ® high throughput immunoassay. J. Clin. Virol. 2021, 136, 104739. [Google Scholar] [CrossRef] [PubMed]
- Demonbreun, A.R.; Sancilio, A.; Velez, M.P.; Ryan, D.T.; Saber, R.; Vaught, L.A.; Reiser, N.L.; Hsieh, R.R.; D’Aquila, R.T.; Mustanski, B.; et al. Comparison of IgG and neutralizing antibody responses after one or two doses of COVID-19 mRNA vaccine in previously infected and uninfected individuals. eClinicalMedicine 2021, 38, 101018. [Google Scholar] [CrossRef]
- Giannella, M.; Huth, M.; Righi, E.; Hasenauer, J.; Marconi, L.; Konnova, A.; Gupta, A.; Hotterbeekx, A.; Berkell, M.; Palacios-Baena, Z.R.; et al. Using machine learning to predict antibody response to SARS-CoV-2 vaccination in solid organ transplant recipients: The multicentre ORCHESTRA cohort. Clin. Microbiol. Infect. 2023, 29, 1084.e1–1084.e7. [Google Scholar] [CrossRef]
- Alejo, J.L.; Mitchell, J.; Chiang, T.P.Y.; Chang, A.; Abedon, A.T.; Werbel, W.A.; William, A.; Boyarsky, B.J.; Zeiser, L.B.; Avery, R.K.; et al. Predicting a Positive Antibody Response After 2 SARS-CoV-2 mRNA Vaccines in Transplant Recipients: A Machine Learning Approach With External Validation. Transplantation 2022, 106, e452–e460. [Google Scholar] [CrossRef] [PubMed]
- Arvin, A.M.; Fink, K.; Schmid, M.A.; Cathcart, A.; Spreafico, R.; Havenar-Daughton, C.; Lanzavecchia, A.; Corti, D.; Virgin, H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature 2020, 584, 353–363. [Google Scholar] [CrossRef]
- Tao, Y.; Bian, S.; Wang, P.; Zhang, H.; Bi, W.; Zhu, P.; Sawan, M. Rapid Optical Biosensing of SARS-CoV-2 Spike Proteins in Artificial Samples. Sensors 2022, 22, 3768. [Google Scholar] [CrossRef]
- Bian, S.; Tao, Y.; Zhu, Z.; Zhu, P.; Wang, Q.; Wu, H.; Sawan, M. On-Site Biolayer Interferometry-Based Biosensing of Carbamazepine in Whole Blood of Epileptic Patients. Biosensors 2021, 11, 516. [Google Scholar] [CrossRef] [PubMed]
- Hentzien, M.; Autran, B.; Piroth, L.; Yazdanpanah, Y.; Calmy, A. A monoclonal antibody stands out against omicron subvariants: A call to action for a wider access to bebtelovimab. Lancet Infect. Dis. 2022, 22, 1278. [Google Scholar] [CrossRef]
- Bian, S.; Berghe, N.V.D.; Vandersmissen, L.; Tops, S.; Vermeire, S.; Ferrante, M.; Gils, A.; Thomas, D. Evaluating an easy sampling method using dried blood spots to determine vedolizumab concentrations. J. Pharm. Biomed. Anal. 2020, 185, 113224. [Google Scholar] [CrossRef]
- Nie, J.; Li, Q.; Wu, J.; Zhao, C.; Hao, H.; Liu, H.; Zhang, L.; Nie, L.; Qin, H.; Wang, M.; et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 2020, 15, 3699–3715. [Google Scholar] [CrossRef]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef]
- Kaushik, A.; Mostafavi, E. To manage long COVID by selective SARS-CoV-2 infection biosensing. Innovation 2022, 3, 100303. [Google Scholar] [CrossRef]
- Yu, X.; Wei, D.; Xu, W.; Li, Y.; Li, X.; Zhang, X.; Qu, J.; Yang, Z.; Chen, E. Reduced sensitivity of SARS-CoV-2 Omicron variant to antibody neutralization elicited by booster vaccination. Cell Discov. 2022, 8, 4. [Google Scholar] [CrossRef]
- Ranzani, O.T.; Hitchings, M.D.T.; de Melo, R.L.; de França, G.V.A.; Fernandes, C.d.F.R.; Lind, M.L.; Torres, M.S.S.; Tsuha, D.H.; David, L.C.S.; Said, R.F.C.; et al. Effectiveness of an inactivated Covid-19 vaccine with homologous and heterologous boosters against Omicron in Brazil. Nat. Commun. 2022, 13, 5536. [Google Scholar] [CrossRef]
- He, C.; Yang, J.; Hong, W.; Chen, Z.; Peng, D.; Lei, H.; Alu, A.; He, X.; Bi, Z.; Jiang, X.; et al. A self-assembled trimeric protein vaccine induces protective immunity against Omicron variant. Nat. Commun. 2022, 13, 5459. [Google Scholar] [CrossRef]
- Subbiah, V. The next generation of evidence-based medicine. Nat. Med. 2023, 29, 49–58. [Google Scholar] [CrossRef]
- Camacho, J.; Albert, E.; Álvarez-Rodríguez, B.; Rusu, L.; Zulaica, J.; Moreno, A.R.; Peiró, S.; Geller, R.; Navarro, D.; Giménez, E. A machine learning model for predicting serum neutralizing activity against Omicron SARS-CoV-2 BA.2 and BA.4/5 sublineages in the general population. J. Med. Virol. 2023, 95, e28739. [Google Scholar] [CrossRef]
- Papadopoulos, D.; Ntanasis-Stathopoulos, I.; Gavriatopoulou, M.; Evangelakou, Z.; Malandrakis, P.; Manola, M.S.; Gianniou, D.D.; Kastritis, E.; Trougakos, I.P.; Dimopoulos, M.A.; et al. Predictive Factors for Neutralizing Antibody Levels Nine Months after Full Vaccination with BNT162b2: Results of a Machine Learning Analysis. Biomedicines 2022, 10, 204. [Google Scholar] [CrossRef]
- Piñana, J.L.; Rodríguez-Belenguer, P.; Caballero, D.; Martino, R.; Lopez-Corral, L.; Terol, M.-J.; Vazquez, L.; Calabuig, M.; Sanz-Linares, G.; Marin-Jimenez, F.; et al. Applicability of probabilistic graphical models for early detection of SARS-CoV-2 reactive antibodies after SARS-CoV-2 vaccination in hematological patients. Ann. Hematol. 2022, 101, 2053–2067. [Google Scholar] [CrossRef] [PubMed]
- Osmanodja, B.; Stegbauer, J.; Kantauskaite, M.; Rump, L.C.; Heinzel, A.; Reindl-Schwaighofer, R.; Oberbauer, R.; Benotmane, I.; Caillard, S.; Masset, C.; et al. Development and validation of multivariable prediction models of serological response to SARS-CoV-2 vaccination in kidney transplant recipients. Front. Immunol. 2022, 13, 997343. [Google Scholar] [CrossRef]
- Lee, W.S.; Tan, H.-X.; Reynaldi, A.; Esterbauer, R.; Koutsakos, M.; Nguyen, J.; Amarasena, T.; Kent, H.E.; Aggarwal, A.; Turville, S.G.; et al. Durable reprogramming of neutralizing antibody responses following Omicron breakthrough infection. Sci. Adv. 2023, 9, eadg5301. [Google Scholar] [CrossRef]
- Shen, X.; Kellogg, R.; Panyard, D.J.; Bararpour, N.; Castillo, K.E.; Lee-McMullen, B.; Delfarah, A.; Ubellacker, J.; Ahadi, S.; Rosenberg-Hasson, Y.; et al. Multi-omics microsampling for the profiling of lifestyle-associated changes in health. Nat. Biomed. Eng. 2023, 8, 11–29. [Google Scholar] [CrossRef] [PubMed]





| No. | Sex | Age Range | Blood Type | BMI | Dose 1, 2, and 3 Vaccines | Interval (Month) | Last Time Point |
|---|---|---|---|---|---|---|---|
| 1 | Female | 21–25 | A | 19.9 | Sinovac | 8.3 | Day 90 |
| 2 | Male | 21–25 | O | 28.7 | Sinopharm | 6.4 | Day 90 |
| 3 | Female | 26–30 | AB | 19.8 | Sinovac | 6.9 | Day 270 |
| 4 | Female | 21–25 | B | 18.6 | Sinovac | 7.0 | Day 90 |
| 5 | Female | 31–35 | O | 24.7 | Sinovac | 5.8 | Day 90 |
| 6 | Female | 21–25 | A | 18.1 | Sinovac | 6.8 | Day 270 |
| 7 | Female | 21–25 | O | 16.8 | Sinovac | 6.1 | Day 270 |
| 8 | Male | 31–35 | B | 21.1 | Sinovac | 7.9 | Day 90 |
| 9 | Female | 21–25 | O | 19.1 | Sinovac | 6.3 | Day 90 |
| 10 | Female | 26–30 | O | 16.6 | Sinovac | 7.2 | Day 270 |
| 11 | Female | 21–25 | O | 20.6 | Sinopharm | 6.5 | Day 90 |
| 12 | Female | 26–30 | O | 19.9 | Sinovac | 9.6 | Day 90 |
| 13 | Female | 26–30 | O | 20.9 | Sinovac | 8.0 | Day 270 |
| 14 | Male | 26–30 | A | 20.1 | Sinovac | 9.0 | Day 270 |
| 15 | Male | 21–25 | AB | 27.5 | Sinovac | 7.7 | Day 90 |
| Input of Previous Measurements (pM), Output at a Given Time Point (T) | Sera NAbs (RMSE) | DBS NAbs (RMSE) | Sera BAbs (RMSE) |
|---|---|---|---|
| 3.1% | 4.2% | 2.7% | |
| 3.7% | 4.0% | 2.6% | |
| 9.0% | 6.3% | 4.2% | |
| 6.0% | 6.5% | 4.7% | |
| 9.9% | 6.1% | 6.5% | |
| 6.6% | 5.3% | 5.3% | |
| 14.4% | 10.1% | 7.8% | |
| 17.3% | 10.1% | 10.2% | |
| 15.9% | 11.2% | 9.5% | |
| 9.4% | 4.9% | 7.1% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bian, S.; Shang, M.; Tao, Y.; Wang, P.; Xu, Y.; Wang, Y.; Shen, Z.; Sawan, M. Dynamic Profiling and Prediction of Antibody Response to SARS-CoV-2 Booster-Inactivated Vaccines by Microsample-Driven Biosensor and Machine Learning. Vaccines 2024, 12, 352. https://doi.org/10.3390/vaccines12040352
Bian S, Shang M, Tao Y, Wang P, Xu Y, Wang Y, Shen Z, Sawan M. Dynamic Profiling and Prediction of Antibody Response to SARS-CoV-2 Booster-Inactivated Vaccines by Microsample-Driven Biosensor and Machine Learning. Vaccines. 2024; 12(4):352. https://doi.org/10.3390/vaccines12040352
Chicago/Turabian StyleBian, Sumin, Min Shang, Ying Tao, Pengbo Wang, Yankun Xu, Yao Wang, Zhida Shen, and Mahamad Sawan. 2024. "Dynamic Profiling and Prediction of Antibody Response to SARS-CoV-2 Booster-Inactivated Vaccines by Microsample-Driven Biosensor and Machine Learning" Vaccines 12, no. 4: 352. https://doi.org/10.3390/vaccines12040352