What are they and how are they useful?
Annotations are biological terms or relations, such as diseases, chemicals or protein interactions, which can be highlighted for readers on abstracts and full text articles. These terms are identified by text mining algorithms, developed by a variety of text mining groups.
For readers, annotations make it easier to scan an article and get a quick overview, find key concepts, and discover evidence, such as gene disease associations or molecular interactions. Annotations enable users to locate the primary data in the text by linking text-mined entities to public life sciences and chemistry databases. The goal of annotations is to support scientists and database curators in their literature research by harnessing the power of text mining, and to promote the contribution of text miners to the advancement of science.
What types of annotations are available?
Annotations are provided for:
- Europe PMC - Database Accessions and Resource names, Gene/Protein names, Organisms, Diseases, Chemicals, Gene Ontology terms and Experimental Methods
- Metagenomics (EMERALD) - Sample-Material, Body-Site, Host, State, Site, Place, Date, Engineered, Ecoregion, Treatment, Kit, Primer, Gene, LS, LCM, Sequencing
- ExTRI - Transcription factors-Gene targets
- IntAct - Protein–protein Interactions
- PubTator - Gene mutations
- PheneBank - Cells, Gene mutations, Phenotypes, Molecules, Anatomy, Pathways
- NaCTEM - Biological events (Phosphorylation)
- OntoGene - Cells, Cell lines, Clinical drugs, Sequences, Molecular processes, Organ/Tissues
- Open Targets Platform and DisGeNET - Gene–Disease associations
- SIB - Gene Function (GeneRIF) statements and COVoc
Using annotations
When viewing an article, a link to 'Annotations' is shown in the tools menu on the right, as shown below.
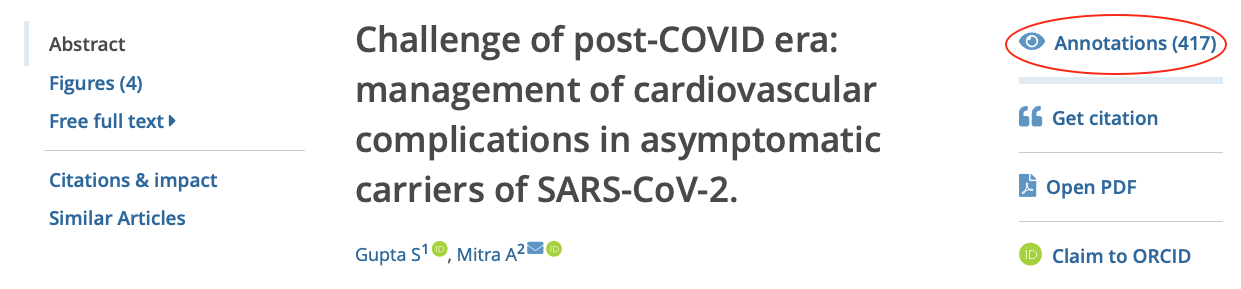
Clicking or tapping on 'Annotations' opens a new panel on the right. Selecting the term in the panel highlights the relevant annotation in the text of the article on the left. Hovering over the highlight opens a popup window.
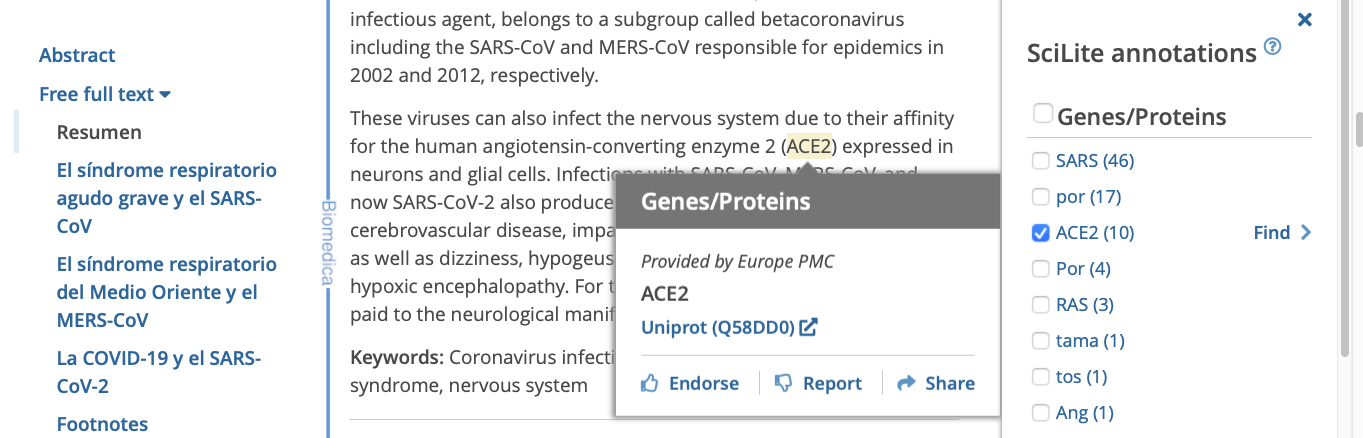
The pop-up window displays a link to related database record, the source of the annotation, and the feedback link. In the case of overlapping annotations in a sentence, we highlight the longest annotation, and the individual annotations within the phrase can be seen in the pop-up window.
Use the 'find' and 'arrow' link to the right of the term to find the first, and subsequent instances of the term in the text.
It is of critical importance that readers find the annotations useful. Readers can provide feedback on each annotation, e.g. mark incorrect annotations or endorse useful ones. This information is fed back to the Europe PMC team and will be acted upon, helping to improve the annotations overall. If you find an incorrect annotation, or the annotation is too generic and is highlighted too often, you can report it by clicking or tapping on the highlighted term and using the Feedback link in the pop-up window. You can also endorse annotations using the Feedback link, if they are useful to you.